DEMO_febio_0043_pyra5_element
Below is a demonstration for:
- Building geometry for a cube with hexahedral elements
- Defining the boundary conditions
- Coding the febio structure
- Running the model
- Importing and visualizing the displacement and stress results
Contents
Keywords
- febio_spec version 4.0
- febio, FEBio
- displacement control, displacement boundary condition
- 5-node linear pyramidal element, pyra5
- pyramid, rectangular
- static, solid
- neo-hookean, uncoupled
- displacement logfile
- stress logfile
clear; close all; clc;
Plot settings
fontSize=20; faceAlpha1=0.5; markerSize=40; markerSize2=35; lineWidth=3; cMap=viridis(20); %colormap cMapMod=gjet(20); %colormap
Control parameters
% Path names defaultFolder = fileparts(fileparts(mfilename('fullpath'))); savePath=fullfile(defaultFolder,'data','temp'); % Defining file names febioFebFileNamePart='tempModel'; febioFebFileName=fullfile(savePath,[febioFebFileNamePart,'.feb']); %FEB file name febioLogFileName=[febioFebFileNamePart,'.txt']; %FEBio log file name febioLogFileName_disp=[febioFebFileNamePart,'_disp_out.txt']; %Log file name for exporting displacement febioLogFileName_stress=[febioFebFileNamePart,'_stress_out.txt']; %Log file name for exporting stress sigma_z febioLogFileName_stretch=[febioFebFileNamePart,'_stretch_out.txt']; %Log file name for exporting stretch U_z febioLogFileName_stress_prin=[febioFebFileNamePart,'_stress_prin_out.txt']; %Log file name for exporting principal stresses febioLogFileName_stretch_prin=[febioFebFileNamePart,'_stretch_prin_out.txt']; %Log file name for exporting principal stretches febioLogFileName_force=[febioFebFileNamePart,'_force_out.txt']; %Log file name for exporting force %Specifying dimensions and number of elements pointSpacing=10; height=10; displacementMagnitude=3; %The displacement magnitude %Material parameter set E_youngs1=0.1; %Material Young's modulus nu1=0.4; %Material Poisson's ratio % FEA control settings numTimeSteps=10; %Number of time steps desired max_refs=25; %Max reforms max_ups=0; %Set to zero to use full-Newton iterations opt_iter=6; %Optimum number of iterations max_retries=5; %Maximum number of retires dtmin=(1/numTimeSteps)/100; %Minimum time step size dtmax=1/numTimeSteps; %Maximum time step size runMode='external';% 'internal' or 'external'
Creating model geometry and mesh
A set of two pyramids is created
% Create a pyramid element V=[pointSpacing/2.*[1,-1,0;1,1,0;-1,1,0;-1,-1,0]; 0 0 pointSpacing/sqrt(2); 0 0 -pointSpacing/sqrt(2)]; E=[1 2 3 4 5; 4 3 2 1 6;]; [F,C,CF]=element2patch(E,(1:1:size(E,1))','pyra5');
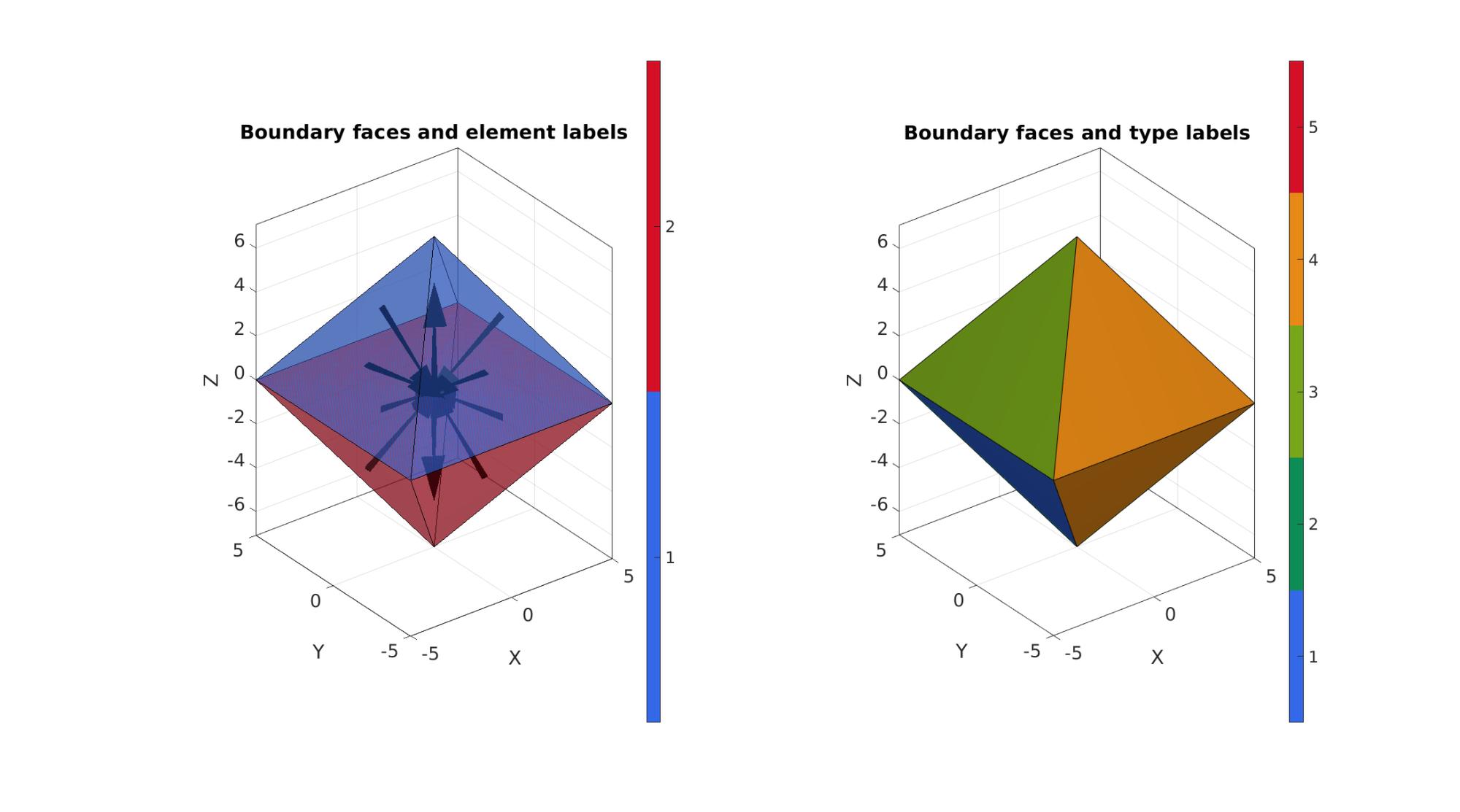
Plotting model boundary surfaces and a cut view
cFigure; subplot(1,2,1); hold on; title('Boundary faces and element labels','FontSize',fontSize); gpatch(F,V,C,'k',faceAlpha1); patchNormPlot(F,V); axisGeom(gca,fontSize); camlight headlight; colormap(gca,cMapMod); icolorbar; subplot(1,2,2); hold on; title('Boundary faces and type labels','FontSize',fontSize); gpatch(F,V,CF,'k',1); axisGeom(gca,fontSize); camlight headlight; colormap(gca,cMapMod); icolorbar; gdrawnow;
Defining the boundary conditions
The visualization of the model boundary shows colors for each side of the cube. These labels can be used to define boundary conditions.
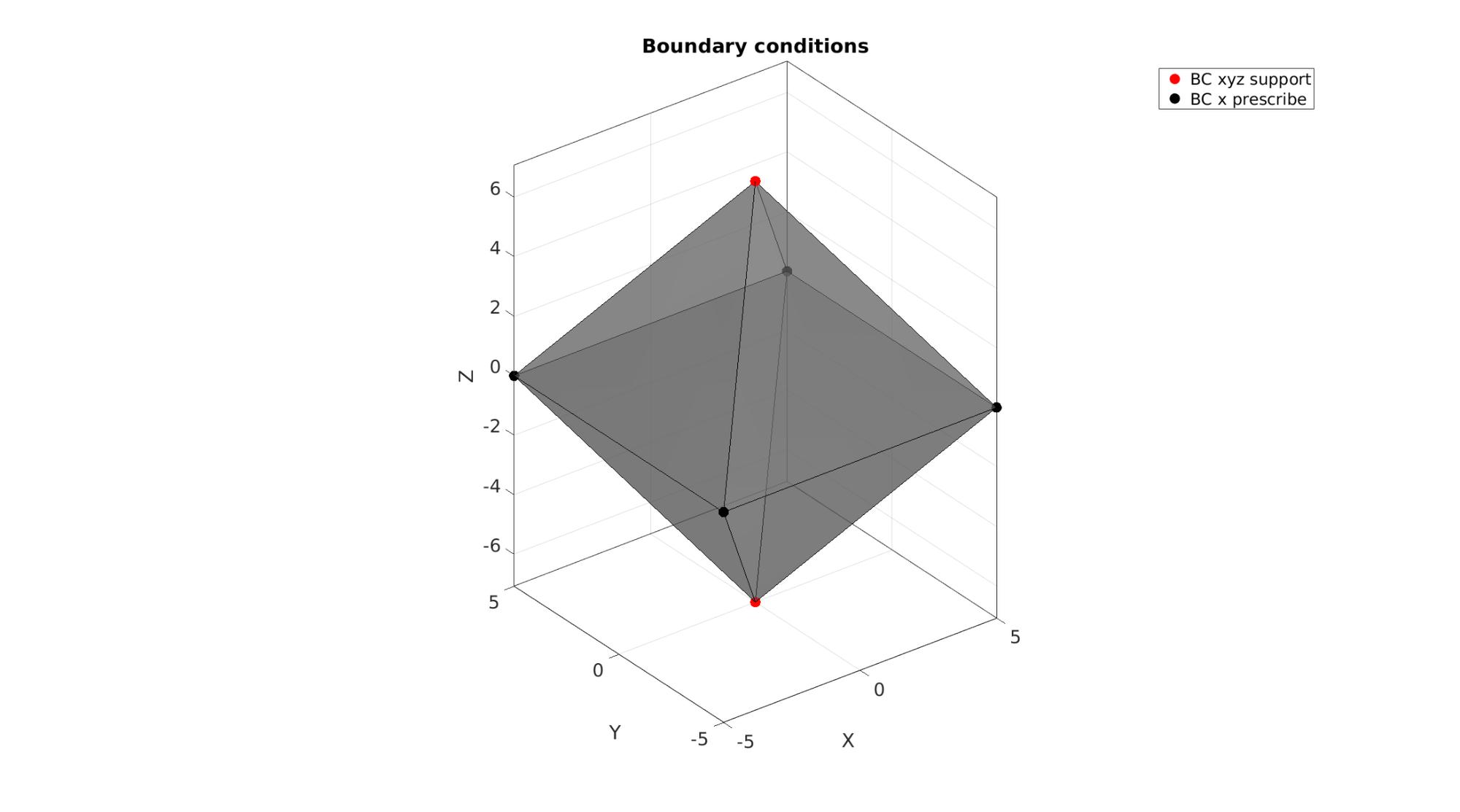
%Define supported node sets bcSupportList=[5 6]; %Prescribed displacement nodes bcPrescribeList=[1 2 3 4];
Visualizing boundary conditions. Markers plotted on the semi-transparent model denote the nodes in the various boundary condition lists.
hf=cFigure; title('Boundary conditions','FontSize',fontSize); xlabel('X','FontSize',fontSize); ylabel('Y','FontSize',fontSize); zlabel('Z','FontSize',fontSize); hold on; gpatch(F,V,'kw','k',0.5); hl(1)=plotV(V(bcSupportList,:),'r.','MarkerSize',markerSize); hl(2)=plotV(V(bcPrescribeList,:),'k.','MarkerSize',markerSize); legend(hl,{'BC xyz support','BC x prescribe'}); axisGeom(gca,fontSize); camlight headlight; drawnow;
Defining the FEBio input structure
See also febioStructTemplate and febioStruct2xml and the FEBio user manual.
%Get a template with default settings [febio_spec]=febioStructTemplate; %febio_spec version febio_spec.ATTR.version='4.0'; %Module section febio_spec.Module.ATTR.type='solid'; %Control section febio_spec.Control.analysis='STATIC'; febio_spec.Control.time_steps=numTimeSteps; febio_spec.Control.step_size=1/numTimeSteps; febio_spec.Control.solver.max_refs=max_refs; febio_spec.Control.solver.qn_method.max_ups=max_ups; febio_spec.Control.time_stepper.dtmin=dtmin; febio_spec.Control.time_stepper.dtmax=dtmax; febio_spec.Control.time_stepper.max_retries=max_retries; febio_spec.Control.time_stepper.opt_iter=opt_iter; %Material section materialName1='Material1'; febio_spec.Material.material{1}.ATTR.name=materialName1; febio_spec.Material.material{1}.ATTR.type='neo-Hookean'; febio_spec.Material.material{1}.ATTR.id=1; febio_spec.Material.material{1}.E=E_youngs1; febio_spec.Material.material{1}.v=nu1; % Mesh section % -> Nodes febio_spec.Mesh.Nodes{1}.ATTR.name='Object1'; %The node set name febio_spec.Mesh.Nodes{1}.node.ATTR.id=(1:size(V,1))'; %The node id's febio_spec.Mesh.Nodes{1}.node.VAL=V; %The nodel coordinates % -> Elements partName1='Part1'; febio_spec.Mesh.Elements{1}.ATTR.name=partName1; %Name of this part febio_spec.Mesh.Elements{1}.ATTR.type='pyra5'; %Element type febio_spec.Mesh.Elements{1}.elem.ATTR.id=(1:1:size(E,1))'; %Element id's febio_spec.Mesh.Elements{1}.elem.VAL=E; %The element matrix % -> NodeSets nodeSetName1='bcSupportList'; nodeSetName2='bcPrescribeList'; febio_spec.Mesh.NodeSet{1}.ATTR.name=nodeSetName1; febio_spec.Mesh.NodeSet{1}.VAL=mrow(bcSupportList); febio_spec.Mesh.NodeSet{2}.ATTR.name=nodeSetName2; febio_spec.Mesh.NodeSet{2}.VAL=mrow(bcPrescribeList); %MeshDomains section febio_spec.MeshDomains.SolidDomain.ATTR.name=partName1; febio_spec.MeshDomains.SolidDomain.ATTR.mat=materialName1; %Boundary condition section % -> Fix boundary conditions febio_spec.Boundary.bc{1}.ATTR.name='zero_displacement_x'; febio_spec.Boundary.bc{1}.ATTR.type='zero displacement'; febio_spec.Boundary.bc{1}.ATTR.node_set=nodeSetName1; febio_spec.Boundary.bc{1}.x_dof=1; febio_spec.Boundary.bc{1}.y_dof=1; febio_spec.Boundary.bc{1}.z_dof=1; febio_spec.Boundary.bc{2}.ATTR.name='zero_displacement_zy'; febio_spec.Boundary.bc{2}.ATTR.type='zero displacement'; febio_spec.Boundary.bc{2}.ATTR.node_set=nodeSetName2; febio_spec.Boundary.bc{2}.x_dof=0; febio_spec.Boundary.bc{2}.y_dof=1; febio_spec.Boundary.bc{2}.z_dof=1; febio_spec.Boundary.bc{3}.ATTR.name='prescibed_displacement_x'; febio_spec.Boundary.bc{3}.ATTR.type='prescribed displacement'; febio_spec.Boundary.bc{3}.ATTR.node_set=nodeSetName2; febio_spec.Boundary.bc{3}.dof='x'; febio_spec.Boundary.bc{3}.value.ATTR.lc=1; febio_spec.Boundary.bc{3}.value.VAL=displacementMagnitude; febio_spec.Boundary.bc{3}.relative=0; %LoadData section % -> load_controller febio_spec.LoadData.load_controller{1}.ATTR.name='LC_1'; febio_spec.LoadData.load_controller{1}.ATTR.id=1; febio_spec.LoadData.load_controller{1}.ATTR.type='loadcurve'; febio_spec.LoadData.load_controller{1}.interpolate='LINEAR'; %febio_spec.LoadData.load_controller{1}.extend='CONSTANT'; febio_spec.LoadData.load_controller{1}.points.pt.VAL=[0 0; 1 1]; %Output section % -> log file febio_spec.Output.logfile.ATTR.file=febioLogFileName; febio_spec.Output.logfile.node_data{1}.ATTR.file=febioLogFileName_disp; febio_spec.Output.logfile.node_data{1}.ATTR.data='ux;uy;uz'; febio_spec.Output.logfile.node_data{1}.ATTR.delim=','; febio_spec.Output.logfile.node_data{2}.ATTR.file=febioLogFileName_force; febio_spec.Output.logfile.node_data{2}.ATTR.data='Rx;Ry;Rz'; febio_spec.Output.logfile.node_data{2}.ATTR.delim=','; febio_spec.Output.logfile.element_data{1}.ATTR.file=febioLogFileName_stress; febio_spec.Output.logfile.element_data{1}.ATTR.data='sz'; febio_spec.Output.logfile.element_data{1}.ATTR.delim=','; febio_spec.Output.logfile.element_data{2}.ATTR.file=febioLogFileName_stretch; febio_spec.Output.logfile.element_data{2}.ATTR.data='Uz'; febio_spec.Output.logfile.element_data{2}.ATTR.delim=','; febio_spec.Output.logfile.element_data{3}.ATTR.file=febioLogFileName_stress_prin; febio_spec.Output.logfile.element_data{3}.ATTR.data='s1;s2;s3'; febio_spec.Output.logfile.element_data{3}.ATTR.delim=','; febio_spec.Output.logfile.element_data{4}.ATTR.file=febioLogFileName_stretch_prin; febio_spec.Output.logfile.element_data{4}.ATTR.data='U1;U2;U3'; febio_spec.Output.logfile.element_data{4}.ATTR.delim=','; % Plotfile section febio_spec.Output.plotfile.compression=0;
Quick viewing of the FEBio input file structure
The febView function can be used to view the xml structure in a MATLAB figure window.
febView(febio_spec); %Viewing the febio file
Exporting the FEBio input file
Exporting the febio_spec structure to an FEBio input file is done using the febioStruct2xml function.
febioStruct2xml(febio_spec,febioFebFileName); %Exporting to file and domNode %system(['gedit ',febioFebFileName,' &']);
Running the FEBio analysis
To run the analysis defined by the created FEBio input file the runMonitorFEBio function is used. The input for this function is a structure defining job settings e.g. the FEBio input file name. The optional output runFlag informs the user if the analysis was run succesfully.
febioAnalysis.run_filename=febioFebFileName; %The input file name febioAnalysis.run_logname=febioLogFileName; %The name for the log file febioAnalysis.disp_on=1; %Display information on the command window febioAnalysis.runMode=runMode; febioAnalysis.maxLogCheckTime=10; %Max log file checking time [runFlag]=runMonitorFEBio(febioAnalysis);%START FEBio NOW!!!!!!!!
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
--------> RUNNING/MONITORING FEBIO JOB <-------- 27-Apr-2023 10:10:28
FEBio path: /home/kevin/FEBioStudio2/bin/febio4
# Attempt removal of existing log files 27-Apr-2023 10:10:28
* Removal succesful 27-Apr-2023 10:10:28
# Attempt removal of existing .xplt files 27-Apr-2023 10:10:28
* Removal succesful 27-Apr-2023 10:10:28
# Starting FEBio... 27-Apr-2023 10:10:28
Max. total analysis time is: Inf s
* Waiting for log file creation 27-Apr-2023 10:10:28
Max. wait time: 10 s
* Log file found. 27-Apr-2023 10:10:28
# Parsing log file... 27-Apr-2023 10:10:28
number of iterations : 1 27-Apr-2023 10:10:28
number of reformations : 1 27-Apr-2023 10:10:28
------- converged at time : 0.1 27-Apr-2023 10:10:28
number of iterations : 1 27-Apr-2023 10:10:28
number of reformations : 1 27-Apr-2023 10:10:28
------- converged at time : 0.2 27-Apr-2023 10:10:28
number of iterations : 1 27-Apr-2023 10:10:28
number of reformations : 1 27-Apr-2023 10:10:28
------- converged at time : 0.3 27-Apr-2023 10:10:28
number of iterations : 1 27-Apr-2023 10:10:28
number of reformations : 1 27-Apr-2023 10:10:28
------- converged at time : 0.4 27-Apr-2023 10:10:28
number of iterations : 1 27-Apr-2023 10:10:28
number of reformations : 1 27-Apr-2023 10:10:28
------- converged at time : 0.5 27-Apr-2023 10:10:28
number of iterations : 1 27-Apr-2023 10:10:28
number of reformations : 1 27-Apr-2023 10:10:28
------- converged at time : 0.6 27-Apr-2023 10:10:28
number of iterations : 1 27-Apr-2023 10:10:28
number of reformations : 1 27-Apr-2023 10:10:28
------- converged at time : 0.7 27-Apr-2023 10:10:28
number of iterations : 1 27-Apr-2023 10:10:28
number of reformations : 1 27-Apr-2023 10:10:28
------- converged at time : 0.8 27-Apr-2023 10:10:28
number of iterations : 1 27-Apr-2023 10:10:28
number of reformations : 1 27-Apr-2023 10:10:28
------- converged at time : 0.9 27-Apr-2023 10:10:28
number of iterations : 1 27-Apr-2023 10:10:28
number of reformations : 1 27-Apr-2023 10:10:28
------- converged at time : 1 27-Apr-2023 10:10:28
Elapsed time : 0:00:00 27-Apr-2023 10:10:28
N O R M A L T E R M I N A T I O N
# Done 27-Apr-2023 10:10:28
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
Import FEBio results
if runFlag==1 %i.e. a succesful run
% Importing nodal displacements from a log file dataStruct=importFEBio_logfile(fullfile(savePath,febioLogFileName_disp),0,1); %Access data N_disp_mat=dataStruct.data; %Displacement timeVec=dataStruct.time; %Time %Create deformed coordinate set V_DEF=N_disp_mat+repmat(V,[1 1 size(N_disp_mat,3)]);
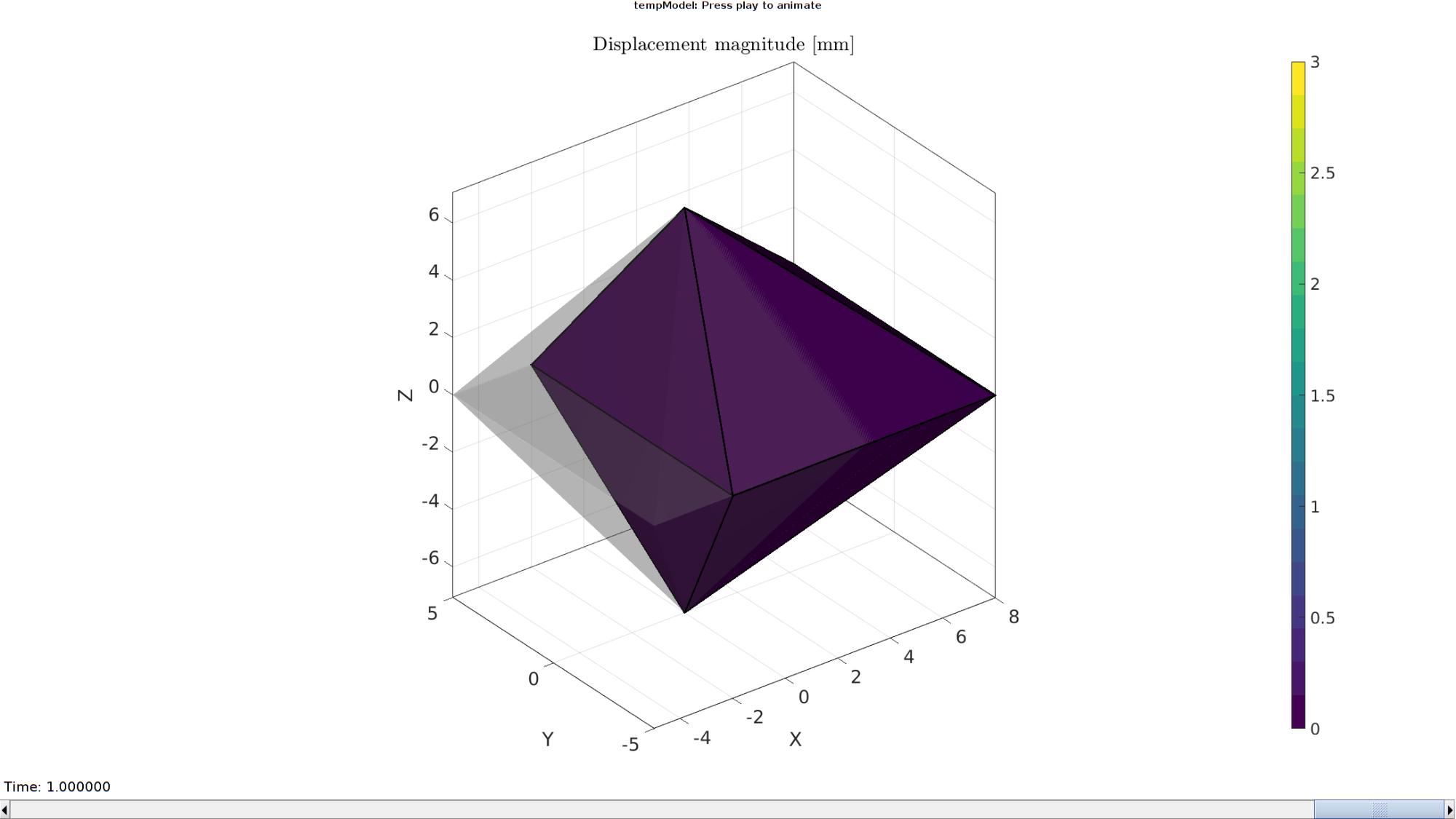
Plotting the simulated results using anim8 to visualize and animate deformations
DN_magnitude=sqrt(sum(N_disp_mat(:,:,end).^2,2)); %Current displacement magnitude % Create basic view and store graphics handle to initiate animation hf=cFigure; %Open figure gtitle([febioFebFileNamePart,': Press play to animate']); title('Displacement magnitude [mm]','Interpreter','Latex') hp=gpatch(F,V_DEF(:,:,end),DN_magnitude,'k',1,2); %Add graphics object to animate gpatch(F,V,0.5*ones(1,3),'none',0.25); %A static graphics object axisGeom(gca,fontSize); colormap(cMap); colorbar; caxis([0 max(DN_magnitude)]); caxis manual; axis(axisLim(V_DEF)); %Set axis limits statically camlight headlight; % Set up animation features animStruct.Time=timeVec; %The time vector for qt=1:1:size(N_disp_mat,3) %Loop over time increments DN_magnitude=sqrt(sum(N_disp_mat(:,:,qt).^2,2)); %Current displacement magnitude %Set entries in animation structure animStruct.Handles{qt}=[hp(1) hp(1) hp(2) hp(2)]; %Handles of objects to animate animStruct.Props{qt}={'Vertices','CData','Vertices','CData'}; %Properties of objects to animate animStruct.Set{qt}={V_DEF(:,:,qt),DN_magnitude,V_DEF(:,:,qt),DN_magnitude}; %Property values for to set in order to animate end anim8(hf,animStruct); %Initiate animation feature drawnow;
end

GIBBON www.gibboncode.org
Kevin Mattheus Moerman, [email protected]
GIBBON footer text
License: https://github.com/gibbonCode/GIBBON/blob/master/LICENSE
GIBBON: The Geometry and Image-based Bioengineering add-On. A toolbox for image segmentation, image-based modeling, meshing, and finite element analysis.
Copyright (C) 2006-2022 Kevin Mattheus Moerman and the GIBBON contributors
This program is free software: you can redistribute it and/or modify it under the terms of the GNU General Public License as published by the Free Software Foundation, either version 3 of the License, or (at your option) any later version.
This program is distributed in the hope that it will be useful, but WITHOUT ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the GNU General Public License for more details.
You should have received a copy of the GNU General Public License along with this program. If not, see http://www.gnu.org/licenses/.