triSurfSelfTriangulateBoundary
Below is a demonstration of the features of the triSurfSelfTriangulateBoundary function
Contents
clear; close all; clc;
Syntax
[F1,V1,ind1]=triSurfSelfTriangulateBoundary(F1,V1,ind1,angleThreshold,isClosedLoop);
Description
The triSurfSelfTriangulateBoundary function
Examples
Example 1: Self-triangulate the boundary of a surface by creating triangles at sharp boundary segments
Create test data set
w=1; [X,Y]=ndgrid(linspace(0,w,15)); Z=ones(size(X)); C=tril(Z); [F,V,C]=surf2patch(X,Y,Z,C); %Quads C=vertexToFaceMeasure(F,C)>0; logicKeep=C>0; F=F(logicKeep,:); C=C(logicKeep,:); [F,V]=patchCleanUnused(F,V); F=[F(:,[1 2 3]);F(:,[3 4 1])]; %Triangles
Get boundary curve
Eb=patchBoundary(F,V);
indBoundaryCurve=edgeListToCurve(Eb);
indBoundaryCurve=indBoundaryCurve(1:end-1)'; %Start=End for closed curve so remove double entry
Self triangulate
angleThreshold=(100/180)*pi; isClosedPath=1; [F_new,V_new,indBoundaryCurve_new]=triSurfSelfTriangulateBoundary(F,V,indBoundaryCurve,angleThreshold,isClosedPath);
cFigure; subplot(1,2,1); hold on; title('Original'); gpatch(F,V,'kw'); plotV(V(indBoundaryCurve,:),'k-','LineWidth',3); axisGeom; view(2); subplot(1,2,2); hold on; title('Self-triangulated'); gpatch(F_new,V_new,'kw'); plotV(V_new(indBoundaryCurve_new,:),'k-','LineWidth',3); axisGeom; view(2); drawnow;
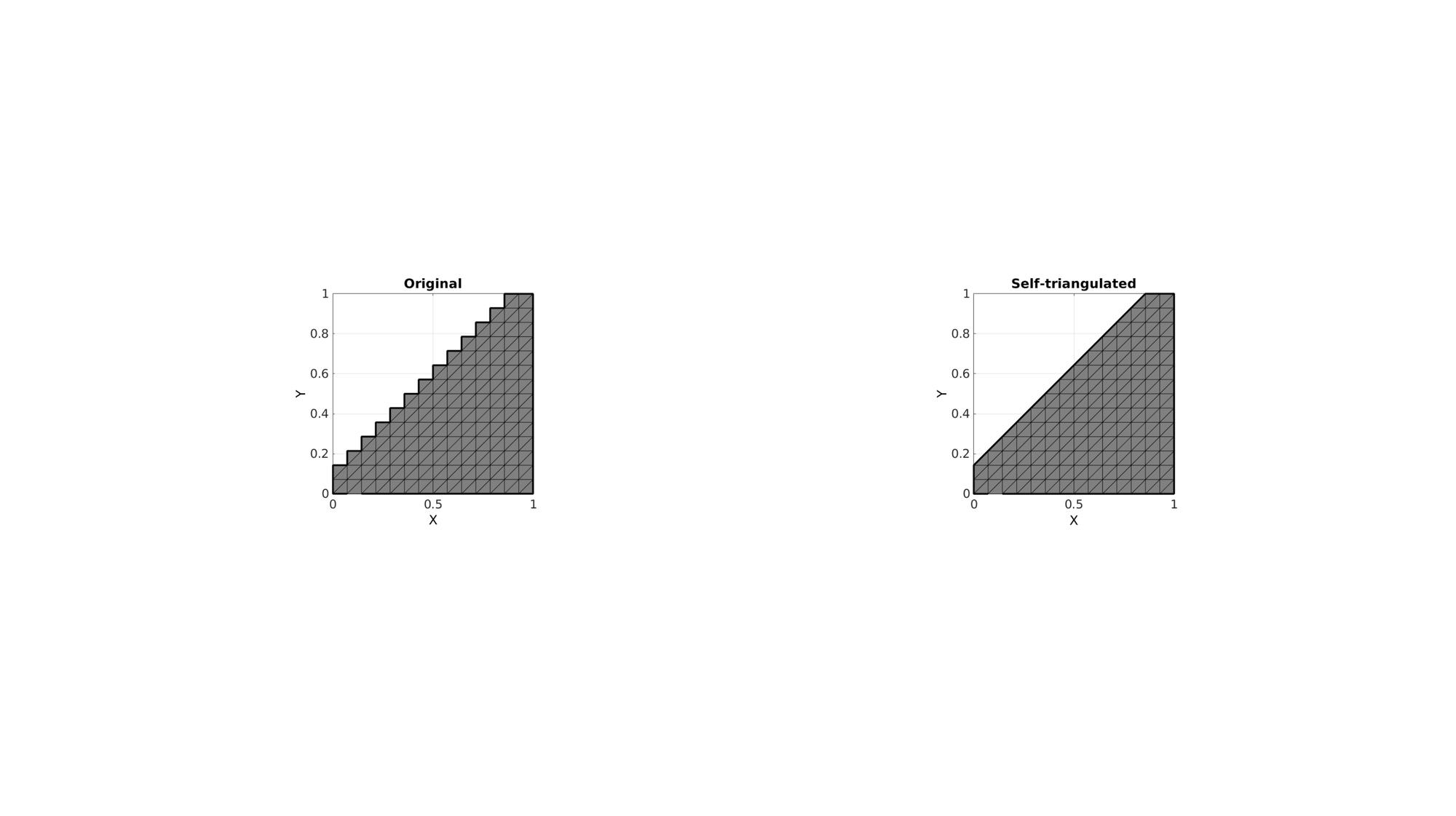
Example 2: Altered shape
V(:,1)=V(:,1)-V(:,2);
Calculate mesh path angles
[F_new,V_new,indBoundaryCurve_new]=triSurfSelfTriangulateBoundary(F,V,indBoundaryCurve,angleThreshold,isClosedPath);
cFigure; subplot(1,2,1); hold on; title('Original'); gpatch(F,V,'kw'); plotV(V(indBoundaryCurve,:),'k-','LineWidth',3); axisGeom; view(2); subplot(1,2,2); hold on; title('Self-triangulated'); gpatch(F_new,V_new,'kw'); plotV(V_new(indBoundaryCurve_new,:),'k-','LineWidth',3); axisGeom; view(2); drawnow;
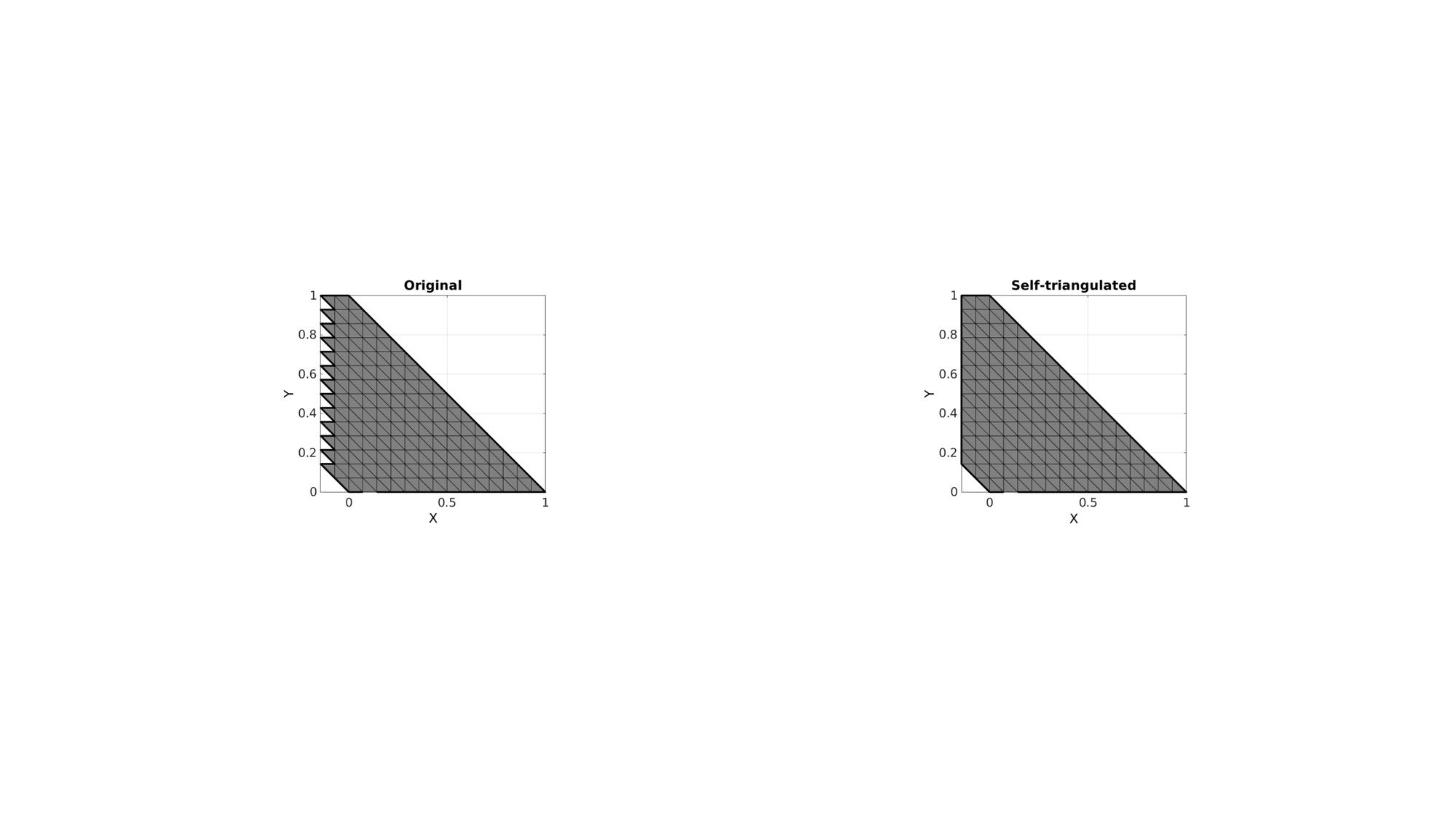
Example 3: Self-triangulate a part of boundary surface
Create path segment
indBoundaryCurve=indBoundaryCurve(1:15);
Calculate mesh path angles
isClosedPath=0; [F_new,V_new,indBoundaryCurve_new]=triSurfSelfTriangulateBoundary(F,V,indBoundaryCurve,angleThreshold,isClosedPath);
cFigure; subplot(1,2,1); hold on; title('Original'); gpatch(F,V,'kw'); plotV(V(indBoundaryCurve,:),'k-','LineWidth',3); axisGeom; view(2); subplot(1,2,2); hold on; title('Self-triangulated'); gpatch(F_new,V_new,'kw'); plotV(V_new(indBoundaryCurve_new,:),'k-','LineWidth',3); axisGeom; view(2); drawnow;
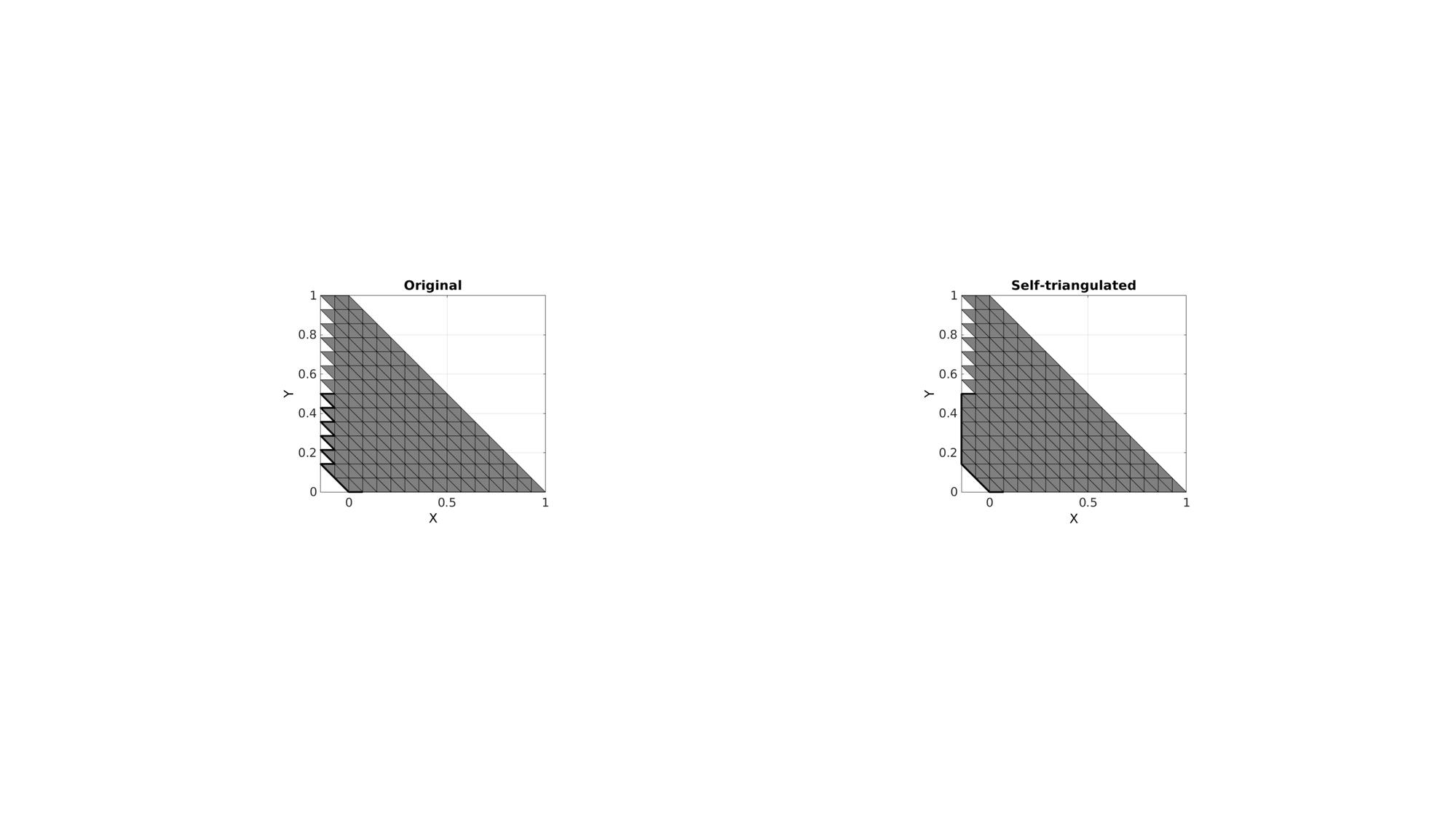

GIBBON www.gibboncode.org
Kevin Mattheus Moerman, [email protected]
GIBBON footer text
License: https://github.com/gibbonCode/GIBBON/blob/master/LICENSE
GIBBON: The Geometry and Image-based Bioengineering add-On. A toolbox for image segmentation, image-based modeling, meshing, and finite element analysis.
Copyright (C) 2019 Kevin Mattheus Moerman
This program is free software: you can redistribute it and/or modify it under the terms of the GNU General Public License as published by the Free Software Foundation, either version 3 of the License, or (at your option) any later version.
This program is distributed in the hope that it will be useful, but WITHOUT ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the GNU General Public License for more details.
You should have received a copy of the GNU General Public License along with this program. If not, see http://www.gnu.org/licenses/.